Research Topics
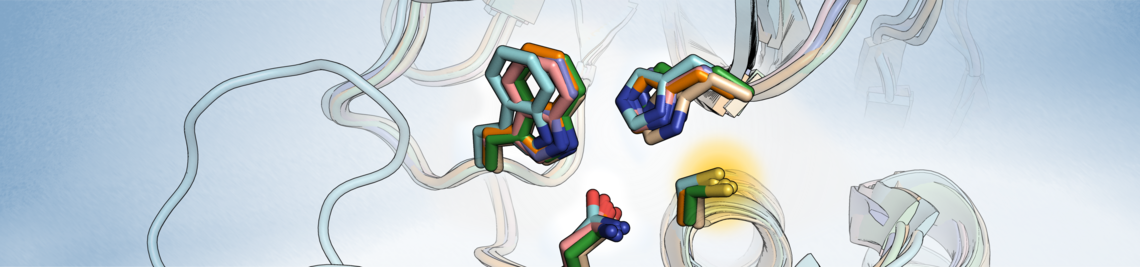
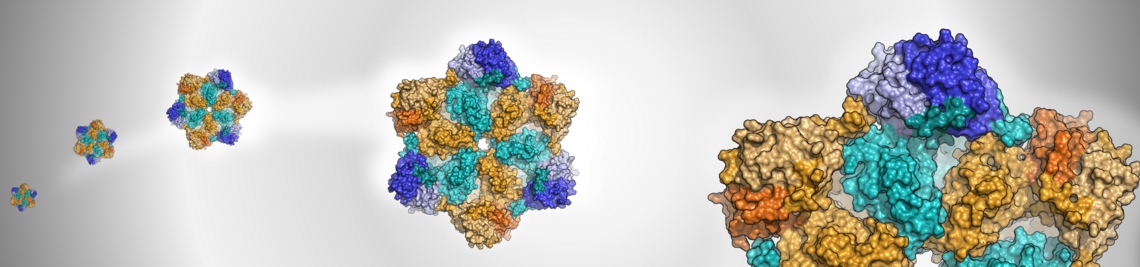
The common theme of our research is the investigation of protein-ligand interactions with computational methods.
In application projects, methods such as docking and molecular dynamics (MD) simulations are used to analyse protein-ligand complexes, to support the interpretation of experimental results, and to establish hypotheses regarding structural and energetic aspects of the interactions. Various approaches of virtual screening are applied to identify new ligands for protein targets of pharmaceutical interest. These projects are frequently embedded in cooperations with research groups in medicinal chemistry, structural biology and pharmacology.
Methods-oriented projects aim to evaluate and advance existing computational methods for drug design. This addresses problems related to affinity prediction, covalent binding, protein flexibility and the treatment of water and solvation effects in the context of protein-ligand interactions. Methods ranging from fast scoring functions to advanced MD simulations and quantum chemical calculations are used in this context.
More about our current research projects, grouped by target, main computational method and overarching topic in drug design, can be found below.