Computation
Docking
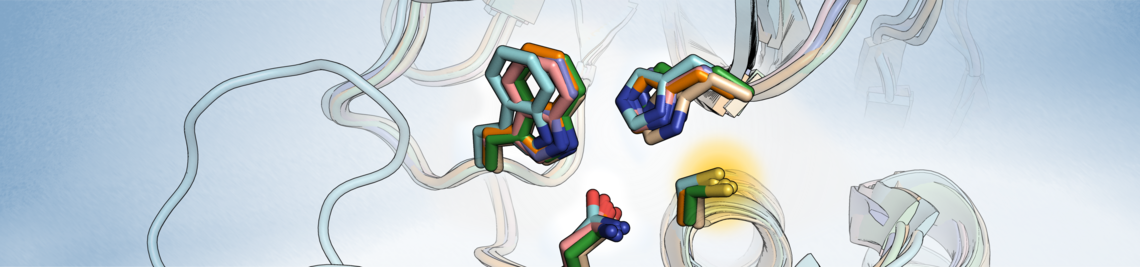
Computational docking is a core method of structure-based drug design, used to predict the binding mode of a ligand to a target protein. We have recently analyzed the performance of docking methods in the context of fragment-based screening. Specific challenges also arise with covalent ligands. The power of docking analyses in providing explanatory models is highlighted by a study where sequence of protein-protein- and protein-ligand-docking allowed to interpret the mode of action of PROTACs on the Aurora-A-kinase.
Publications on this topic:
C. Herbst, S. Endres, R. Würz, C. Sotriffer
Assessment of fragment docking and scoring with the endothiapepsin model system
Arch. Pharm. 2024, 357 (6), e2400061
B. Adhikari, J. Bozilovic, M. Diebold, J. D. Schwarz, J. Hofstetter, M. Schröder, M. Wanior, A. Narain, M. Vogt, N. D. Stankovic, A. Baluapuri, L. Schönemann, L. Eing, P. Bhandare, B. Kuster, A. Schlosser, S. Heinzlmeir, C. Sotriffer, S. Knapp, E. Wolf
PROTAC-mediated degradation reveals a non-catalytic function of AURORA-A kinase
Nat. Chem. Biol. 2020, 16, 1179-1188
C. Sotriffer
Docking of covalent ligands: challenges and approaches
Molecular Informatics 2018, 37, 1800062
Scoring functions
Scoring functions are used in docking and virtual screening to assess the quality of binding modes, to rank different ligands and to estimate binding affinities. We have worked in the past on empirical scoring functions and have developed one of the first machine-learning based scoring functions, called SFCscoreRF.
Publications on this topic:
L. Pason, C.Sotriffer
Empirical scoring functions for affinity prediction of protein-ligand complexes
Molecular Informatics 2016, 35, 541-548
D. Zilian, C. Sotriffer
SFCscoreRF: A random-forest-based scoring function for improved affinity prediction of protein-ligand complexes
J. Chem. Inf. Model. 2013, 53, 1923-1933
Virtual screening
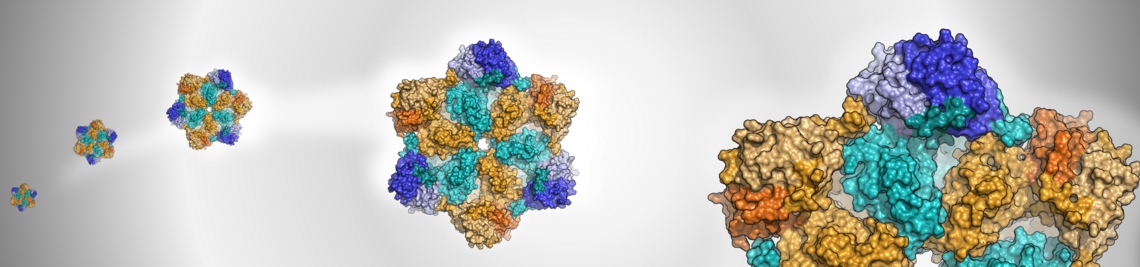
Virtual screening is a computational workflow to identify biocactive molecules with a desired property profile from large compound libraries. Many different approaches and techniques are used for this purpose, as presented in a book edited by us some years ago. An example of how to screen for selective inhibitors of a particular target is illustrated by a study on N-myristoyltransferases, in which we collaborated with the Brenk lab at the University of Bergen.
Publications on this topic:
C. Kersten, E. Fleischer, J. Kehrein, C. Borek, E. Jaenicke, C. Sotriffer, R. Brenk
How to design selective ligands for highly conserved binding sites: A case study using N-myristoyltransferases as a model system
J. Med. Chem. 2020, 63, 2095-2113
C. Sotriffer (editor)
Virtual Screening: Principles, Challenges, and Practical Guidelines
Wiley‐VCH, 2011
Molecular dynamics simulations
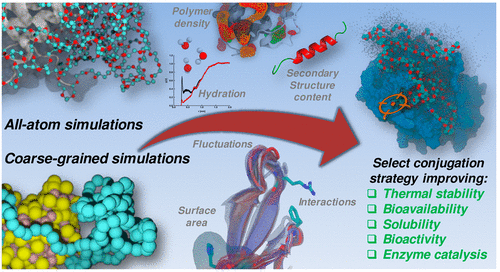
MD simulations are essential to capture the effects of protein flexibility and solvation in structure-based drug design. Two recent studies in which we applied MD simulations for rationalizing experimental inhibition data are listed below. A further field of application are polymers, as illustrated by a review on simulations for polymer bioconjugation. Polymer MD studies from our group can be found in the full publication list.
Publications on this topic:
T. Lohr, C. Herbst, N. M. Bzdyl, C. Jenkins, N. J. Scheuplein, W. O. Sugiarto, J. J. Whittaker, A. Guskov, I. Norville, U. A. Hellmich, F. Hausch, M. Sarkar-Tyson, C. Sotriffer, U. Holzgrabe
High affinity inhibitors of the macrophage infectivity potentiator protein from Trypanosoma cruzi, Burkholderia pseudomallei, and Legionella pneumophila - A comparison
ACS Infect. Dis. 2024, 10, 3681-3691
J. V. Patzke, F. Sauer, R. K. Nair, E. Endres, E. Proschak, V. Hernandez-Olmos, C. Sotriffer, C. Kisker
Structural basis for the bi-specificity of USP25 and USP28 inhibitors
EMBO Rep. 2024, 25, 2950-2973
J. Kehrein, C. Sotriffer
Molecular dynamics simulations for rationalizing polymer bioconjugation strategies: Challenges, recent developments, and future opportunities
ACS Biomater. Sci. Eng. 2024, 10, 51-74